The Unix Shell
Opening
Nelle Nemo, a marine biologist, has just returned from a six-month survey of the North Pacific Gyre, where she has been sampling gelatinous marine life in the Great Pacific Garbage Patch. She has 1520 samples in all, and now needs to:
- Run each sample through an assay machine that will measure the relative abundance of 300 different proteins. The machine's output for a single sample is a file with one line for each protein.
-
Calculate statistics for each of the proteins separately
using a program her supervisor wrote called
goostat. -
Compare the statistics for each protein with corresponding statistics for each other protein
using a program one of the other graduate students wrote called
goodiff. - Write up. Her supervisor would really like her to do this by the end of the month so that her paper can appear in an upcoming special issue of Aquatic Goo Letters.
It takes about half an hour for the assay machine to process each sample. The good news is, it only takes two minutes to set each one up. Since her lab has eight assay machines that she can use in parallel, this step will "only" take about two weeks.
The bad news is,
if she has to run goostat and goodiff by hand,
she'll have to enter filenames and click "OK" roughly 3002 times
(300 runs of goostat, plus 300×299 runs of goodiff).
At 30 seconds each,
that will 750 hours, or 18 weeks.
Not only would she miss her paper deadline,
the chances of her getting all 90,000 commands right are approximately zero.
This chapter is about what she should do instead. More specifically, it's about how she can use a command shell to automate the repetitive steps in her processing pipeline, so that her computer can work 24 hours a day while she writes her paper. As a bonus, once she has put a processing pipeline together, she will be able to use it again whenever she collects more data.
Loops
Objectives
- Write a loop that applies one or more commands separately to each file in a set of files.
- Trace the values taken on by a loop variable during execution of the loop.
- Explain the difference between a variable's name and its value.
- Explain why spaces and some punctuation characters shouldn't be used in files' names.
- Demonstrate how to see what commands have recently been executed.
- Re-run recently executed commands without retyping them.
Duration: 15-20 minutes.
Lesson
Wildcards and tabs are one way to save on typing.
Another is to tell the shell to do something over and over again.
Suppose we have several hundred genome data files in a directory
with names like basilisk.dat,
unicorn.dat,
and so on.
When new files arrive,
we'd like to rename the existing ones to
original-basilisk.dat,
original-unicorn.dat,
etc.
We can't use:
mv *.dat original-*.dat
because that would expand (in the two-file case) to:
mv basilisk.dat unicorn.dat
This wouldn't back up our files:
it would replace the content of unicorn.dat
with whatever's in basilisk.dat.
Instead, we can use a loop to do some operation once for each thing in a list. Here's a simple example that displays the first three lines of each file in turn:
$ for filename in basilisk.dat unicorn.dat > do > head -3 $filename > done COMMON NAME: basilisk CLASSIFICATION: basiliscus vulgaris UPDATED: 1745-05-02 COMMON NAME: unicorn CLASSIFICATION: equus monoceros UPDATED: 1738-11-24
When the shell sees the keyword for,
it knows it is supposed to repeat a command (or group of commands)
once for each thing in a list.
In this case,
the list is the two filenames.
Each time through the loop,
the name of the thing currently being operated on
is assigned to the variable filename.
Inside the loop,
we get the variable's value by putting $ in front of it
(this is known as shell parameter expansion),
so the first time through the loop,
$filename is basilisk.dat,
and the second time,
unicorn.dat.
Finally,
the command that's actually being run is our old friend head,
so this loop prints out the first three lines of each data file in turn.
Follow the Prompt
You may have noticed that the shell prompt changed
from $ to > and back again
as we were typing in our loop.
The second prompt, >,
is different to remind us that we haven't finished typing a complete command yet.
What's In a Name?
We have called the variable in this loop filename
in order to make its purpose clearer to human readers.
The shell itself doesn't care what the variable is called
(but when we use the variable value it is a good practice to use
${filename} instead of $filename to avoid
some types of problems);
if we wrote this loop as:
for x in basilisk.dat unicorn.dat
do
head -3 $x
done
or:
for temperature in basilisk.dat unicorn.dat
do
head -3 $temperature
done
it would work exactly the same way.
Don't do this.
Programs are only useful if people can understand them,
so using meaningless names (like x)
or misleading names (like temperature)
increases the likelihood of the program being wrong.
Here's a slightly more complicated loop:
for filename in *.dat
do
echo $filename
head -100 $filename | tail -20
done
The shell starts by expanding *.dat
to create the list of files it will process.
The loop body then executes two commands
for each of those files.
The first,
echo,
just prints its command-line parameters to standard output.
For example:
echo hello there
prints:
hello there
In this case,
since the shell expands $filename to be the name of a file,
echo $filename just prints the name of the file.
Note that we can't write this as:
for filename in *.dat
do
$filename
head -100 $filename | tail -20
done
because then the first time through the loop,
when $filename expanded to basilisk.dat,
the shell would try to run basilisk.dat as a program.
Finally,
the head and tail combination
selects lines 80-100 from whatever file is being processed.
Spaces in Names
Filename expansion in loops is another reason you should not use spaces in filenames. Suppose our data files are named:
basilisk.dat red dragon.dat unicorn.dat
If we try to process them using:
for filename in *.dat
do
echo $filename
head -100 $filename | tail -20
done
then *.dat will expand to:
basilisk.dat red dragon.dat unicorn.dat
which means that filename will be assigned
each of the following values in turn:
basilisk.dat
red
dragon.dat
unicorn.dat
The highlighted lines show the problem:
instead of getting one name red dragon.dat,
the commands in the loop will get red and dragon.dat separately.
To make matters worse,
the file red dragon.dat won't be processed at all.
There are ways to get around this, the simplest of which is to put quote marks around "$filename",
but the safest thing is to use dashes,
underscores,
or some other printable character instead of spaces in your file names.
Going back to our original file renaming problem, we can solve it using this loop:
for filename in *.dat
do
mv $filename original-$filename
done
This loop runs the mv command once for each filename.
The first time,
when $filename expands to basilisk.dat,
the shell executes:
mv basilisk.dat original-basilisk.dat
The second time, the command is:
mv unicorn.dat original-unicorn.dat
Measure Twice, Run Once
A loop is a way to do many things at once—or to make many mistakes at once if it does the wrong thing. One way to check what a loop would do is to echo the commands it would run instead of actually running them. For example, we could write our file renaming loop like this:
for filename in *.dat
do
echo mv $filename original-$filename
done
Instead of running mv,
this loop runs echo,
which prints out:
mv basilisk.dat original-basilisk.dat mv unicorn.dat original-unicorn.dat
without actually running those commands.
We can then use up-arrow to redisplay the loop,
back-arrow to get to the word echo,
delete it,
and then press "enter" to run the loop
with the actual mv commands.
This isn't foolproof,
but it's a handy way to see what's going to happen
when you're still learning how loops work.
So far we have been renaming files of the form someanimal.dat to
original-someanimal.dat. But, what if instead of
original-someanimal.dat we want
someanimal.dat.original or someanimal-original.dat?
For someanimal.dat.original we can do
for filename in *.dat
do
mv $filename $filename.original
done
But for someanimal-original.dat we will need to go a little deeper
in shell parameter expansion and use its find and replace feature. That syntax
is ${variable/find/substitute/} where find is the
longest string in variable value that we want to be replaced by
substitute. We can achieve this task using
for filename in *.dat
do
mv $filename ${filename/.dat/-original.dat/}
done
Nelle's Pipeline: Processing Files
Nelle is now ready to process her data files. Since she's still learning how to use the shell, she decides to build up the required commands in stages. Her first step is to make sure that she can select the right files—remember, these are ones whose names end in 'A' or 'B', rather than 'Z':
$ cd north-pacific-gyre/2012-07-03 $ for datafile in *[AB].txt do echo $datafile done NENE01729A.txt NENE01729B.txt NENE01736A.txt ... NENE02043A.txt NENE02043B.txt $
Her next step is to figure out what to call the files
that the goostat analysis program will create.
Prefixing each input file's name with "stats" seems simple,
so she modifies her loop to do that:
$ for datafile in *[AB].txt do echo $datafile stats-$datafile done NENE01729A.txt stats-NENE01729A.txt NENE01729B.txt stats-NENE01729B.txt NENE01736A.txt stats-NENE01736A.txt ... NENE02043A.txt stats-NENE02043A.txt NENE02043B.txt stats-NENE02043B.txt $
She hasn't actually run goostats yet,
but now she's sure she can select the right files
and generate the right output filenames.
Typing in commands over and over again is becoming tedious, though, and Nelle is worried about making mistakes, so instead of re-entering her loop, she presses the up arrow. In response, Bash redisplays the whole loop on one line (using semi-colons to separate the pieces):
$ for datafile in *[AB].txt; do echo $datafile stats-$datafile; done
Using the left arrow key,
Nelle backs up and changes the command echo to goostats:
$ for datafile in *[AB].txt; do goostats $datafile stats-$datafile; done
When she presses enter, Bash runs the modified command. However, nothing appears to happen—there is no output. After a moment, Nelle realizes that since her script doesn't print anything to the screen any longer, she has no idea whether it is running, much less how quickly. She kills the job by typing Control-C, uses up-arrow to repeat the command, and edits it to read:
$ for datafile in *[AB].txt; do echo $datafile; goostats $datafile stats-$datafile; done
When she runs her program now, it produces one line of output every five seconds or so:
NENE01729A.txt
NENE01729B.txt
NENE01736A.txt
...
$
1518 times 5 seconds, divided by 60,
tells her that her script will take about two hours to run.
As a final check,
she opens another terminal window,
goes into north-pacific-gyre/2012-07-03,
and uses cat NENE01729B.txt to examine
one of the output files.
It looks good,
so she decides to get some coffee and catch up on her reading.
Those Who Know History Can Choose to Repeat It
Another way to repeat previous work is to use the history command
to get a list of the last few hundred commands that have been executed,
and then to use !123 (where "123" is replaced by the command number)
to repeat one of those commands.
For example,
if Nelle types this:
$ $ history | tail -5 456 ls -l NENE0*.txt 457 rm stats-NENE01729B.txt.txt 458 goostats NENE01729B.txt stats-NENE01729B.txt 459 ls -l NENE0*.txt 460 history
then she can re-run goostats on NENE01729B.txt
simply by typing !458.
Summary
- Use a
forloop to repeat commands once for every thing in a list. - Every
forloop needs a variable to refer to the current "thing". - Use
$nameto expand a variable (i.e., get its value). - Do not use spaces, quotes, or wildcard characters such as '*' or '?' in filenames, as it complicates variable expansion.
- Give files consistent names that are easy to match with wildcard patterns to make it easy to select them for looping.
- Use the up-arrow key to scroll up through previous commands to edit and repeat them.
- Use
historyto display recent commands, and!numberto repeat a command by number.
Challenges
Suppose that
lsinitially displays:fructose.dat glucose.dat sucrose.datWhat is the output of:
for datafile in *.dat do ls *.dat doneIn the same directory, what is the effect of this loop?
for sugar in *.dat do echo $sugar cat $sugar > xylose.dat done-
Prints
fructose.dat,glucose.dat, andsucrose.dat, and copiessucrose.datto createxylose.dat. -
Prints
fructose.dat,glucose.dat, andsucrose.dat, and concatenates all three files to createxylose.dat. -
Prints
fructose.dat,glucose.dat,sucrose.dat, andxylose.dat, and copiessucrose.datto createxylose.dat. - None of the above.
-
Prints
The
exprdoes simple arithmetic using command-line parameters:$ expr 3 + 5 8 $ expr 30 / 5 - 2 4
Given this, what is the output of:
for left in 2 3 do for right in $left do expr $left + $right done doneDescribe in words what the following loop does.
for how in frog11 prcb redig do $how -limit 0.01 NENE01729B.txt doneThe loop:
for num in {1..3} do echo $num doneprints:
1 2 3However, the loop:
for num in {0.1..0.3} do echo $num doneprints:
{0.1..0.3}Write a loop that prints:
0.1 0.2 0.3
Shell Scripts
Objectives
- Write a shell script that runs a command or series of commands for a fixed set of files.
- Run a shell script from the command line.
- Write a shell script that operates on a set of files defined by the user on the command line.
- Create pipelines that include user-written shell scripts.
Duration: 15 minutes.
Lesson
We are finally ready to see what makes the shell such a powerful programming environment. We are going to take the commands we repeat frequently and save them in files so that we can re-run all those operations again later by typing a single command. For historical reasons, a bunch of commands saved in a file is usually called a shell script, but make no mistake: these are actually small programs.
Let's start by putting the following line in the file middle.sh:
head -20 cholesterol.pdb | tail -5
This is a variation on the pipe we constructed earlier:
it selects lines 15-20 of the file cholesterol.pdb.
Remember,
we are not running it as a command just yet:
we are putting the commands in a file.
Text vs. Whatever
We usually call programs like Microsoft Word or LibreOffice Writer "text editors"
(a more correct name is word processor),
but we need to be a bit more careful when it comes to programming.
By default,
Microsoft Word uses .doc (and LibreOffice Writer uses
.odt) files to store not only text,
but also formatting information about fonts,
headings,
and so on.
This extra information isn't stored as characters,
and doesn't mean anything to the shell interpreter:
it expects input files to contain nothing but the letters, digits, and punctuation
on a standard computer keyboard.
When editing programs,
therefore,
you must either use a plain text editor,
or be careful to save files as plain text.
Once we have saved the file,
we can ask the shell to execute the commands it contains.
Our shell is called bash,
so we run the following command:
$ bash middle.sh ATOM 14 C 1 -1.463 -0.666 1.001 1.00 0.00 ATOM 15 C 1 0.762 -0.929 0.295 1.00 0.00 ATOM 16 C 1 0.771 -0.937 1.840 1.00 0.00 ATOM 17 C 1 -0.664 -0.610 2.293 1.00 0.00 ATOM 18 C 1 -4.705 2.108 -0.396 1.00 0.00
Sure enough, our script's output is exactly what we would get if we ran that pipeline directly.
Running Scripts Without Calling Bash
We actually don't need to call Bash explicitly when we want to run a shell script. Instead, we can change the permissions on the shell script so that the operating system knows it is runnable, then put a special line at the start to tell the OS what program to use to run it. for an explanation of how to do this.
What if we want to select lines from an arbitrary file?
We could edit middle.sh each time to change the filename,
but that would probably take longer than just retyping the command.
Instead,
let's edit middle.sh
and replace cholesterol.pdb with a special variable called $1:
$ cat middle.sh head -20 $1 | tail -5
Inside a shell script,
$1 means "the first filename (or other parameter) on the command line".
We can now run our script like this:
$ bash middle.sh cholesterol.pdb ATOM 14 C 1 -1.463 -0.666 1.001 1.00 0.00 ATOM 15 C 1 0.762 -0.929 0.295 1.00 0.00 ATOM 16 C 1 0.771 -0.937 1.840 1.00 0.00 ATOM 17 C 1 -0.664 -0.610 2.293 1.00 0.00 ATOM 18 C 1 -4.705 2.108 -0.396 1.00 0.00
or on a different file like this:
$ bash middle.sh vitamin_a.pdb ATOM 14 C 1 1.788 -0.987 -0.861 ATOM 15 C 1 2.994 -0.265 -0.829 ATOM 16 C 1 4.237 -0.901 -1.024 ATOM 17 C 1 5.406 -0.117 -1.087 ATOM 18 C 1 -0.696 -2.628 -0.641
We still need to edit middle.sh each time we want to adjust the range of lines, though.
Let's fix that by using the special variables $2 and $3:
$ cat middle.sh head $2 $1 | tail $3 $ bash middle.sh vitamin_a.pdb -20 -5 ATOM 14 C 1 1.788 -0.987 -0.861 ATOM 15 C 1 2.994 -0.265 -0.829 ATOM 16 C 1 4.237 -0.901 -1.024 ATOM 17 C 1 5.406 -0.117 -1.087 ATOM 18 C 1 -0.696 -2.628 -0.641
What if we want to process many files in a single pipeline? For example, if we want to sort our PDB files by length, we would type:
$ wc -l *.pdb | sort -n
because wc -l lists the number of lines in the files,
and sort -n sorts things numerically.
We could put this in a file,
but then it would only ever sort a list of PDB files in the current directory.
If we want to be able to get a sorted list of other kinds of files,
possibly from many sub-directories,
we need a way to get all those names into the script.
We can't use $1, $2, and so on
because we don't know how many files there are.
Instead,
we use the special variable $*,
which means, "All of the command-line parameters to the shell script."
Here's an example:
$ cat sorted.sh wc -l $* | sort -n $ bash sorted.sh *.dat backup/*.dat 29 chloratin.dat 89 backup/chloratin.dat 91 sphagnoi.dat 156 sphag2.dat 172 backup/sphag-merged.dat 182 girmanis.dat
Why Isn't It Doing Anything?
What happens if a script is supposed to process a bunch of files, but we don't give it any filenames? For example, what if we type:
$ bash sorted.sh
but don't say *.dat (or anything else)?
In this case,
$* expands to nothing at all,
so the pipeline inside the script is effectively:
wc -l | sort -n
Since it doesn't have any filenames,
wc assumes it is supposed to process standard input,
so it just sits there and waits for us to give it some data interactively.
From the outside,
though,
all we see is it sitting there:
the script doesn't appear to do anything.
But now consider a script called distinct.sh:
sort $* | uniq
Let's run it as part of a pipeline without any filenames:
$ head -5 *.dat | bash distinct.sh
This is equivalent to:
$ head -5 *.dat | sort | uniq
which is actually what we want.
We have two more things to do before we're finished with our simple shell scripts. If you look at a script like:
wc -l $* | sort -n
you can probably puzzle out what it does. On the other hand, if you look at this script:
# List files sorted by number of lines.
wc -l $* | sort -n
you don't have to puzzle it out—the comment at the top of the script tells you what it does. A line or two of documentation like this make it much easier for other people (including your future self) to re-use your work. The only caveat is that each time you modify the script, you should check that the comment is still accurate: an explanation that sends the reader in the wrong direction is worse than none at all.
Second, suppose we have just run a series of commands that did something useful—for example, that created a graph we'd like to use in a paper. We'd like to be able to re-create the graph later if we need to, so we want to save the commands in a file. Instead of typing them in again (and potentially getting them wrong), we can do this:
$ history | tail -4 > redo-figure-3.sh
The file redo-figure-3.sh now contains:
297 goostats -r NENE01729B.txt stats-NENE01729B.txt 298 goodiff stats-NENE01729B.txt /data/validated/01729.txt > 01729-differences.txt 299 cut -d ',' -f 2-3 01729-differences.txt > 01729-time-series.txt 300 ygraph --format scatter --color bw --borders none 01729-time-series.txt figure-3.png
After a moment's work in an editor to remove the serial numbers on the command, we have a completely accurate record of how we created that figure.
Unnumbering
Nelle could also use colrm
(short for "column removal")
to remove the serial numbers on her previous commands.
In practice, most people develop shell scripts by running commands at the shell prompt a few times to make sure they're doing something useful, and doing it correctly, then saving them in a file for re-use. This style of work allows people to recycle what they discover about their data and their workflow with just a few extra keystrokes.
Nelle's Pipeline: Creating a Script
An off-hand comment from her supervisor has made Nelle realize that
she should have provided a couple of extra parameters to goostats
when she processed her files.
This might have been a disaster if she had done all the analysis by hand,
but thanks to for loops,
it will only take a couple of hours to re-do.
Experience has taught her, though, that if something needs to be done twice, it will probably need to be done a third or fourth time as well. She runs the editor and writes the following:
# Calculate reduced stats for data files at J = 100 c/bp.
for datafile in $*
do
echo $datafile
goostats -J 100 -r $datafile stats-$datafile
done
(The parameters -J 100 and -r
are the ones her supervisor said she should have used.)
She saves this in a file called do-stats.sh,
so that she can now re-do the first stage of her analysis by typing:
$ bash do-stats.sh *[AB].txt
She can also do this:
$ bash do-stats.sh *[AB].txt | wc -l
so that the output is just the number of files processed, rather than the names of the files that were processed.
One thing to note about Nelle's script is her choice to let the person running it decide what files to process. She could have written the script as:
# Calculate reduced stats for A and Site B data files at J = 100 c/bp.
for datafile in *[AB].txt
do
echo $datafile
goostats -J 100 -r $datafile stats-$datafile
done
The advantage is that this always selects the right files:
she doesn't have to remember to exclude the 'Z' files.
The disadvantage is that it always selects just those files—she
can't run it on all files (including the 'Z' files),
or on the 'G' or 'H' files her colleagues in Antarctica are producing,
without editing the script.
If she wanted to be more adventurous,
she could modify her script to check for command-line parameters,
and use *[AB].txt if none were provided.
Of course,
this introduces another tradeoff between flexibility and complexity;
we'll explore this later.
Summary
- Save commands in files (usually called shell scripts) for re-use.
- Use
bash filenameto run saved commands. $*refers to all of a shell script's command-line parameters.$1,$2, etc., refer to specified command-line parameters.- Letting users decide what files to process is more flexible and more consistent with built-in Unix commands.
Challenges
Leah has several hundred data files, each of which is formatted like this:
2013-11-05,deer,5 2013-11-05,rabbit,22 2013-11-05,raccoon,7 2013-11-06,rabbit,19 2013-11-06,deer,2 2013-11-06,fox,1 2013-11-07,rabbit,18 2013-11-07,bear,1
Write a shell script called
species.shthat takes any number of filenames as command-line parameters, and usescut,sort, anduniqto print a list of the unique species appearing in each of those files separately.Write a shell script called
latest.shthat takes the name of a directory and a filename extension as its parameters, and prints out the name of the most recently modified file in that directory with that extension. For example:$ bash latest.sh /tmp/data pdbwould print the name of the PDB file in
/tmp/datathat has been changed most recently.If you run the command:
history | tail -5 > recent.sh
the last command in the file is the
historycommand itself, i.e., the shell has addedhistoryto the command log before actually running it. In fact, the shell always adds commands to the log before running them. Why do you think it does this?Joel's
datadirectory contains three files:fructose.dat,glucose.dat, andsucrose.dat. Explain what each of the following shell scripts does when run asbash scriptname *.dat.1. echo *.*
2. for filename in $1 $2 $3 do cat $filename done3. echo $*.dat
Finding Things
Objectives
- Use
grepto select lines from text files that match simple patterns. - Use
findto find files whose names match simple patterns. - Use the output of one command as the command-line parameters to another command.
- Explain what is meant by "text" and "binary" files, and why many common tools don't handle the latter well.
Duration: 15 minutes.
Lesson

You can often guess someone's age by listening to how people talk about search (Figure 17). Just as young people use "Google" as a verb, crusty old Unix programmers use "grep". The word is a contraction of "global/regular expression/print", a common sequence of operations in early Unix text editors. It is also the name of a very useful command-line program.
grep finds and prints lines in files that match a pattern.
For our examples,
we will use a file that contains three haikus
taken from a 1998 competition in Salon magazine:
The Tao that is seen Is not the true Tao, until You bring fresh toner. With searching comes loss and the presence of absence: "My Thesis" not found. Yesterday it worked Today it is not working Software is like that.
Let's find lines that contain the word "not":
$ grep not haiku.txt Is not the true Tao, until "My Thesis" not found Today it is not working $
Here, not is the pattern we're searching for.
It's pretty simple: every alphanumeric character matches against itself.
After the pattern comes the name or names of the files we're searching in.
The output is the three lines in the file that contain the letters "not".
Let's try a different pattern: "day".
$ grep day haiku.txt Yesterday it worked Today it is not working $
This time,
the output is lines containing the words "Yesterday" and "Today",
which both have the letters "day".
If we give grep the -w flag,
it restricts matches to word boundaries,
so that only lines with the word "day" will be printed:
$ grep -w day haiku.txt
$
In this case, there aren't any, so grep's output is empty.
Another useful option is -n, which numbers the lines that match:
$ grep -n it haiku.txt 5:With searching comes loss 9:Yesterday it worked 10:Today it is not working $
Here, we can see that lines 5, 9, and 10 contain the letters "it".
As with other Unix commands, we can combine flags.
For example,
since -i makes matching case-insensitive,
and -v inverts the match,
using them both only prints lines that don't match the pattern
in any mix of upper and lower case:
$ grep -i -v the haiku.txt You bring fresh toner. With searching comes loss Yesterday it worked Today it is not working Software is like that. $
grep has lots of other options.
To find out what they are, we can type man grep.
man is the Unix "manual" command.
It prints a description of a command and its options,
and (if you're lucky) provides a few examples of how to use it:
$ man grep GREP(1) GREP(1) NAME grep, egrep, fgrep - print lines matching a pattern SYNOPSIS grep [OPTIONS] PATTERN [FILE...] grep [OPTIONS] [-e PATTERN | -f FILE] [FILE...] DESCRIPTION grep searches the named input FILEs (or standard input if no files are named, or if a single hyphen- minus (-) is given as file name) for lines containing a match to the given PATTERN. By default, grep prints the matching lines. … … … OPTIONS Generic Program Information --help Print a usage message briefly summarizing these command-line options and the bug-reporting address, then exit. -V, --version Print the version number of grep to the standard output stream. This version number should be included in all bug reports (see below). Matcher Selection -E, --extended-regexp Interpret PATTERN as an extended regular expression (ERE, see below). (-E is specified by POSIX.) -F, --fixed-strings Interpret PATTERN as a list of fixed strings, separated by newlines, any of which is to be matched. (-F is specified by POSIX.) … … …
Wildcards
grep's real power doesn't come from its options, though;
it comes from the fact that patterns can include wildcards.
(The technical name for these is
regular expressions,
which is what the "re" in "grep" stands for.)
Regular expressions are complex enough that
we devoted an entire section of the website to them;
if you want to do complex searches,
please check it out.
As a taster,
we can find lines that have an 'o' in the second position like this:
$ grep -E '^.o' haiku.txt
You bring fresh toner.
Today it is not working
Software is like that.
We use the -E flag and put the pattern in quotes to prevent the shell from trying to interpret it.
(If the pattern contained a '*', for example, the shell would try to expand it before running grep.)
The '^' in the pattern anchors the match to the start of the line.
The '.' matches a single character
(just like '?' in the shell),
while the 'o' matches an actual 'o'.
While grep finds lines in files,
the find command finds files themselves.
Again, it has a lot of options;
to show how the simplest ones work, we'll use the directory tree in
Figure 18:
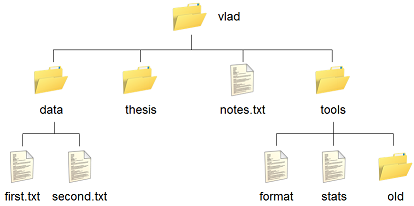
Vlad's home directory contains one file called notes.txt and four subdirectories:
thesis (which is sadly empty),
data (which contains two files first.txt and second.txt),
a tools directory that contains the programs format and stats,
and an empty subdirectory called old.
For our first command, let's run find . -type d.
. is the directory where we want our search to start;
-type d means "things that are directories".
Sure enough,
find's output is the names of the five directories in our little tree
(including ., the current working directory):
$ find . -type d ./ ./data ./thesis ./tools ./tools/old $
If we change -type d to -type f,
we get a listing of all the files instead:
$ find . -type f ./data/first.txt ./data/second.txt ./notes.txt ./tools/format ./tools/stats $
find automatically goes into subdirectories,
their subdirectories,
and so on to find everything that matches the pattern we've given it.
If we don't want it to,
we can use -maxdepth to restrict the depth of search:
$ find . -maxdepth 1 -type f ./notes.txt $
The opposite of -maxdepth is -mindepth,
which tells find to only report things that are at or below a certain depth.
-mindepth 2 therefore finds all the files that are two or more levels below us:
$ find . -mindepth 2 -type f ./data/first.txt ./data/second.txt ./tools/format ./tools/stats $
Another option is -empty,
which restricts matches to empty files and directories:
$ find . -empty ./thesis ./tools/old $
Now let's try matching by name:
$ find . -name *.txt ./notes.txt $
We expected it to find all the text files,
but it only prints out ./notes.txt.
The problem is that the shell expands wildcard characters like * before commands run.
Since *.txt in the current directory expands to notes.txt,
the command we actually ran was:
$ find . -name notes.txt
find did what we asked; we just asked for the wrong thing.
To get what we want,
let's do what we did with grep:
put *.txt in single quotes to prevent the shell from expanding the * wildcard.
This way,
find actually gets the pattern *.txt,
not the expanded filename notes.txt:
$ find . -name '*.txt' ./data/first.txt ./data/second.txt ./notes.txt $
As we said earlier,
the command line's power lies in combining tools.
We've seen how to do that with pipes; let's look at another technique.
As we just saw, find . -name '*.txt' gives us a list of all text files in or below the current directory.
How can we combine that with wc -l to count the lines in all those files?
One way is to put the find command inside $():
$ wc -l $(find . -name '*.txt') 70 ./data/first.txt 420 ./data/second.txt 30 ./notes.txt 520 total $
When the shell executes this command,
the first thing it does is run whatever is inside the $().
It then replaces the $() expression with that command's output.
Since the output of find is the three filenames
./data/first.txt, ./data/second.txt, and ./notes.txt,
the shell constructs the command:
$ wc -l ./data/first.txt ./data/second.txt ./notes.txt
which is what we wanted.
This expansion is exactly what the shell does when it expands wildcards like * and ?,
but lets us use any command we want as our own "wildcard".
It's very common to use find and grep together.
The first finds files that match a pattern;
the second looks for lines inside those files that match another pattern.
Here, for example, we can find PDB files that contain iron atoms
by looking for the string "FE" in all the .pdb files below the current directory:
$ grep FE $(find . -name '*.pdb') ./human/heme.pdb:ATOM 25 FE 1 -0.924 0.535 -0.518 $
Binary Files
We have focused exclusively on finding things in text files.
What if your data is stored as images, in databases, or in some other format?
One option would be to extend tools like grep to handle those formats.
This hasn't happened, and probably won't, because there are too many formats to support.
The second option is to convert the data to text,
or extract the text-ish bits from the data.
This is probably the most common approach,
since it only requires people to build one tool per data format (to extract information).
On the one hand, it makes simple things easy to do.
On the negative side, complex things are usually impossible.
For example,
it's easy enough to write a program that will extract X and Y dimensions from image files for grep to play with,
but how would you write something to find values in a spreadsheet whose cells contained formulas?
The third choice is to recognize that the shell and text processing have their limits, and to use a programming language such as Python instead. When the time comes to do this, don't be too hard on the shell: many modern programming languages, Python included, have borrowed a lot of ideas from it, and imitation is also the sincerest form of praise.
Nelle's Pipeline: The Second Stage
Nelle now has a directory called north-pacific-gyre/2012-07-03
containing 1518 data files,
and needs to compare each one against all of the others
to find the hundred pairs with the highest pairwise scores.
Armed with what she has learned so far,
she writes the following script
# Compare all pairs of files.
for left in $*
do
for right in $*
do
echo $left $right $(goodiff $left $right)
done
done
The outermost loop assigns the name of each file to the variable left in turn.
The inner loop does the same thing for the variable right
each time the outer loop executes,
so inside the inner loop,
left and right are given
each pair of filenames.
Each time it runs the command inside the inner loop,
the shell starts by running goodiff on the two files
in order to expand the $() expression.
Once it's done that,
it passes the output,
along with the names of the files,
to echo.
Thus,
if Nelle saves this script as pairwise.sh
and runs it as:
$ bash pairwise.sh stats-*.txt
the shell runs:
echo stats-NENE01729A.txt stats-NENE01729A.txt $(goodiff stats-NENE01729A.txt stats-NENE01729A.txt) echo stats-NENE01729A.txt stats-NENE01729B.txt $(goodiff stats-NENE01729A.txt stats-NENE01729B.txt) echo stats-NENE01729A.txt stats-NENE01736A.txt $(goodiff stats-NENE01729A.txt stats-NENE01736A.txt) ...
which turns into:
echo stats-NENE01729A.txt stats-NENE01729A.txt files are identical echo stats-NENE01729A.txt stats-NENE01729B.txt 0.97182 echo stats-NENE01729A.txt stats-NENE01736A.txt 0.45223 ...
which in turn prints:
stats-NENE01729A.txt stats-NENE01729A.txt files are identical stats-NENE01729A.txt stats-NENE01729B.txt 0.97182 stats-NENE01729A.txt stats-NENE01736A.txt 0.45223 ...
That's a good start, but Nelle can do better. First, she notices that when the two input files are the same, the output is the words "files are identical" instead of a numerical score. She can remove these lines like this:
$ bash pairwise.sh stats-*.txt | grep -v 'files are identical'
or put the call to grep inside the shell script
(which will be less error-prone):
for left in $*
do
for right in $*
do
echo $left $right $(goodiff $left $right)
done
done | grep -v 'files are identical'
This works because do…done
counts as a single command in Bash.
If Nelle wanted to make this clearer,
she could put parentheses around the loop:
(for left in $* do for right in $* do echo $left $right $(goodiff $left $right) done done) | grep -v 'files are identical'
The last thing Nelle needs to do before writing up
is find the 100 best pairwise matches.
She has seen this before:
sort the lines numerically,
then use head to select the top lines.
However,
the numbers she wants to sort by are at the end of the line,
rather than beginning.
Reading the output of man sort tells her
that the -k flag will let her specify
which column of input she wants to use as a sort key,
but the syntax looks a little complicated.
Instead,
she moves the score to the front of each line:
(for left in $*
do
for right in $*
do
echo $(goodiff $left $right) $left $right
done
done) | grep -v 'files are identical'
and then adds two more commands to the pipeline:
(for left in $*
do
for right in $*
do
echo $(goodiff $left $right) $left $right
done
done) | grep -v 'files are identical' | sort -n -r | head -100
Since this is hard to read,
she uses \ to tell the shell
that she's continuing commands on the next line:
(for left in $*
do
for right in $*
do
echo $(goodiff $left $right) $left $right
done
done) \
| grep -v 'files are identical' \
| sort -n -r \
| head -100
She then runs:
$ bash pairwise.sh stats-*.txt > top100.txt
and heads off to a seminar,
confident that by the time she comes back tomorrow,
top100.txt will contain
the results she needs for her paper.
Summary
- Everything is stored as bytes, but the bytes in binary files do not represent characters.
- Use nested loops to run commands for every combination of two lists of things.
- Use
\to break one logical line into several physical lines. - Use parentheses
()to keep things combined. - Use
$(command)to insert a command's output in place. findfinds files with specific properties that match patterns.grepselects lines in files that match patterns.man commanddisplays the manual page for a given command.
Challenges
The
middle.shscript's numerical parameters are the number of lines to take from the head of the file, and the number of lines to take from the tail of that range. It would be more intuitive if the parameters were the index of the first line to keep and the number of lines, e.g., if:$ bash middle.sh somefile 50 5selected lines 50-54 instead of lines 46-50. Use
$()and theexprcommand to do this.Write a short explanatory comment for the following shell script:
find . -name '*.dat' -print | wc -l | sort -n
The
-vflag togrepinverts pattern matching, so that only lines which do not match the pattern are printed. Given that, which of the following commands will find all files in/datawhose names end inose.dat(e.g.,sucrose.datormaltose.dat), but do not contain the wordtemp?1. find /data -name '*.dat' -print | grep ose | grep -v temp
2. find /data -name ose.dat -print | grep -v temp
3. grep -v temp $(find /data -name '*ose.dat' -print)
4. None of the above. Nelle has saved the IDs of some suspicious observations in a file called
maybe-faked.txt:NENE01909C NEMA04220A NEMA04301A
Which of the following scripts will search all of the
.txtfiles whose names begin withNEfor references to these records?1. for pat in maybe-faked.txt do find . -name 'NE*.txt' -print | grep $pat done2. for pat in $(cat maybe-faked.txt) do grep $pat $(find . -name 'NE*.txt' -print) done3. for pat in $(cat maybe-faked.txt) do find . -name 'NE*.txt' -print | grep $pat done4.. None of the above.
Summary
The Unix shell is older than most of the people who use it. It has survived so long because it is one of the most productive programming environments ever created—maybe even the most productive. Its syntax may be cryptic, but people who have mastered it can experiment with different commands interactively, then use what they have learned to automate their work. Graphical user interfaces may be better at the first, but the shell is still unbeaten at the second. And as Alfred North Whitehead wrote in 1911, "Civilization advances by extending the number of important operations which we can perform without thinking about them."
